Isolation and Molecular Characterization of Salmonella spp. from Beef.
Author: Ferdous Hasan Mithun*
Introduction:
Innumberable foodborne zoonotic pathogen, Salmonella pose a serious threat to human health and food safety. It may present in the intestinal tract of chicken, cattle, swine, poultry and so on. For Salmonella isolation at least 25 gm of sample is required. Afterwards,it is necessary to culture in specific culture media and for farther confirmation PCR may required.
 |
| Salmonella spp. |
General Characteristics:
Morphology:
Family: Enterobacteriaceae
Gram-negative rods
Motile except Salmonella gallinarum and S. pullorum
Aerobic and facultatively anaerobic
Non-spore former, non-capsulated.
 |
| Salmonella characteristics |
Cultural Characteristics:
Grow on ordinary culture media( Nutrient agar)
Aerobic/ Facultatively anaerobic
Temp. 15-41℃
Colonies appear as large 2-3 mm, circular, low convex
On MacConkey medium appear colorless ( NLF)
Selective medium – Wilson Blair Bismath Sulphide medium. Produce Jet black colonies H₂S produced by Salmonella typhi
 |
| Chemical Characteristics |
Biochemical Characteristics:
Do not ferment lactose or sucrose
Do not produce indole
Ferment glucose, mannitol, maltose with production of acid and gas except S.typhi (produces acid only)
Most strain produces H₂S in TSI agar except S.paratyphi and S. choleraesuis
Methyl red : Positive
 |
| Biochemical Characteristics |
Materials and Methods:
Isolation and Identification of Salmonella spp.
Bacterial genomic DNA extraction
Identification of Salmonella spp. by PCR
 |
| Meet sample |
Materials:
Sample: Beef
Media: Salmonella-Shigella agar (SS), EMB agar, XLD agar and MH agar
Nutrient broth, Peptone broth, MR-VP and Sugar media.
Diluents: 0.1% Peptone water.
Homogenizer: Colwarth Stomacher blender
Others: Test tube, Petridishes, Conical Flask, Pipettes, Slides, Coverslips, Incubator, Freeze,Water bath,Microscope,Glass spreader, Gel-Electrophoresis apparatus etc.
Sample Collection:
The sample (Beef-500gm) was collected from commercial local market, Kewatkhali, Mymensingh and it was transported to the laboratory of the Dept. of Microbiology and Hygiene, BAU.
The sample was used to isolate and identify and also to characterized Salmonellae by biochemical,serological and molecular methods.
Isolation of Salmonella organism from Beef:
Step-1. 50 gm sample was taken from the collected sample
Step-2: Added diluent ( 0.1% Peptone water-450ml)
Step-3: Made homogenous mixture by Colwarth Stomacher blender and taken to the test tube( original sample)
Step-4: A series of test tubes, each containing 9ml of 0.1% Peptone water were taken
Step-5: 1ml transferred in test tube no.1 and mixed thoroughly
Step-6: From 1st tube, 1ml transferred to the 2nd tube and continued to the last tube and finally 1 ml discarded
Step-7: For each tube 3 petridishes taken which contain SS agar
Step-8: 0.5ml of mixture transferred from test tube to the petridishes
Step-9: After marking kept in the incubator ( 37℃for overnight)
Step-10: Pure black colony observed, indicates Salmonella spp.
Identification:
1. Pure black colony, subcultured on SS agar
2. Gram’s staining
3. Gram ( - ) ve , Small rod shaped bacteria
4. Subcultured on to XLD agar
5. Incubation at 37℃ at overnight
6. Small, circular pink colony with black center
7. It’s Salmonellae
Media Colony:
 |
| Salmonella colony on SS agar |
 |
| Cultural Characteristics on EMB agar |
Gram Staining:
 |
| Gram Staining |
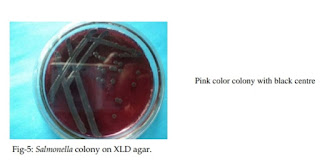 |
| Colony on XLD agar |
 |
| Fig. Biochemical Characteristics |
Biochemical Characteristics:
Molecular Characterization by PCR:
DNA Extraction
A pure bacterial colony was mixed with 100µl of distilled water
Boiled for 10 mins.
immediately kept on ice for ice shock
Finally centrifugation was done at 10,000 rpm for 10 mins.
The supernatant were collected and used as DNA template for PCR
Primer Used for RCR:
 |
| Fig. Primer |
Identification of Salmonella by PCR:
Genus specific PCR was performed to amplify inVA gene
Two set primers( SpeF and fimA) were used for this purpose
Each 20µl reaction mixture consist of 3µl genomic DNA, 10µl PCR master mixtures , 1µl of each of the two primers with the final volume adjusted to 20µl with 5µl of nuclease free water.
Amplification was done by PCR:
Initial denaturation at 94℃ for 5 mins.
Followed by 20 cycles of denaturation at 94℃ for 1 mins.
Annealing temp. of primer was 58℃ for 45 sec.
Elongation at 70℃ for 1 min
Final elongation at 72℃ for 2 mins.
The final holding temp. was 4℃
1% and 3% agarose gel were used for electrophoresis of SpeF & fimA gene respectively.
 |
Fig.PCR for Salmonella gallinarum ( Lane1 was 1 kb ladder,Lane2,5 were negative for SpeF gene ,Lane3,4 were positive and 6 was negative control) |
 |
Fig. PCR for Salmonella typhimurium detecting fimA gene. (Lane1:100 by ladder,Lane2,3,4 were positive and 5,6,7 were negative and 8 was negative control |
References:
Md. Shafiullah Parvej,June-2013.Isolation,identification and molecular characterization of Beef Salmonellae.Reaserchgate.net/publication/297563780
Ahmed AM,Shimabukuro H,Shimamoto T,2009.Isolation and molecular characterization of multidrug resistant strains of E.coli and Salmonella from reatail chicken meat in Japan. J.Food Sci.74:405
Jawad AAK, Hamadani AHA, 2011. Detection of fimA & fimC genes of Salmonella isolates using Polymerase Chain Reaction. J.of Basrah Reseaches(Sciences).37(4):1817-2695




No comments:
Post a Comment
Never share any links or personal information. Keep your privacy strong.